|
 |
| Tuberc Respir Dis > Volume 76(2); 2014 > Article |
|
Abstract
Background
Variable-number tandem repeat (VNTR) typing is a promising method to discriminate the Mycobacterium tuberculosis isolates in molecular epidemiology. The purpose of this study is to determine the optimal VNTR combinations for discriminating isolated M. tuberculosis strains in Korea.
Methods
A total of 317 clinical isolates collected throughout Korea were genotyped by using the IS6110 restriction fragment length polymorphism (RFLP), and then analysed for the number of VNTR copies from 32 VNTR loci.
Results
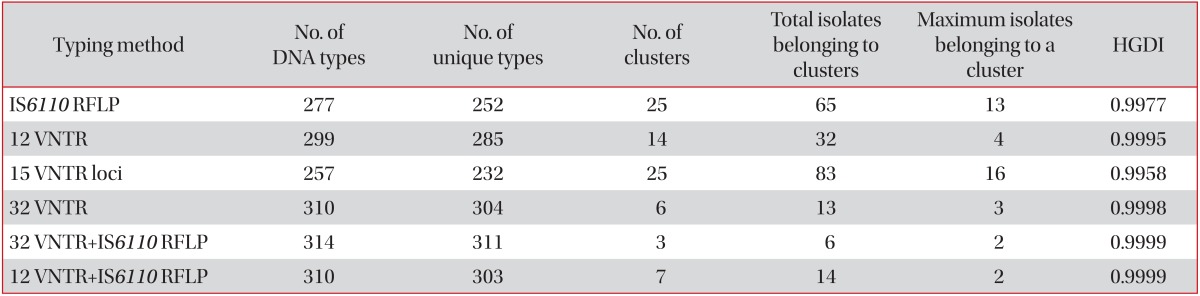
The results of discriminatory power according to diverse combinations were as follows: 25 clusters in 83 strains were yielded from the internationally standardized 15 VNTR loci (Hunter-Gaston discriminatory index [HGDI], 0.9958), 25 clusters in 65 strains by using IS6110 RFLP (HGDI, 0.9977), 14 clusters in 32 strains in 12 hyper-variable VNTR loci (HGDI, 0.9995), 6 clusters in 13 strains in 32 VNTR loci (HDGI, 0.9998), and 7 clusters in 14 strains of both the 12 hyper-variable VNTR and IS6110 RFLP (HDGI, 0.9999).
Tuberculosis (TB) is a contagious disease that develops from infection with Mycobacterium tuberculosis bacilli in droplets projected by coughing of active TB patients. DNA typing of M. tuberculosis is an efficient tool to show the transmission link of TB1. In particular, IS6110 restriction fragment length polymorphism (RFLP) has been used for discrimination of Korean M. tuberculosis isolates because of high IS6110 copies and diverse patterns2-4. However, IS6110 RFLP has some disadvantages such as a lengthy turnaround time, difficulty in comparison between laboratory DNA typing data, and uncertain discrimination of isolates containing similar fragments sizes for inter- or intra-strains5.
Among more than 40 variable-number tandem repeat (VNTR) loci scattered on the M. tuberculosis chromosome6, 15 and 24 VNTR loci have been proposed as the international standard7. However, the discriminatory power is not enough in countries that have a high proportion of Beijing type M. tuberculosis8-12. K strains that have 10 IS6110 RFLP copies are most dominant strain and often found in TB outbreak in Korea. K strains occupy about 4-5% out of any group of M. tuberculosis isolates isolated in Korea3,13. K strains causes difficulty in discerning the direct transmission link by IS6110 RFLP typing only.
Therefore, we attempted to optimize a combination of VNTR loci and IS6110 RFLP for discrimination of Korean M. tuberculosis isolates.
A total of 317 strains were randomly selected among 2,400 strains isolated from smear and culture-positive primary pulmonary TB patients that registered at the Public Health Center (PHC) of Korea between 2006 and 2011. When the strains were clustered in IS6110 RFLP or VNTR typing, we collected the epidemiological information of the strains from the PHCs and each person who had the clustered strain by documents and telephone calls.
For all 317 isolates, DNA isolation and IS6110 RFLP typing were performed as described previously2. An RFLP cluster was defined by completely identical patterns after analysis of BioNumerics version 5.1 software (Applied Maths, Kortrjk, Belgium) within two or more isolates. K strains were identified according to the previously reported definition4,13.
We selected 32 VNTR loci for this study7,9. Primer sets for polymerase chain reaction (PCR) were prepared for the 32 VNTR loci. PCR was conducted as described previously14-16 and VNTR 4052, 3155, and 2074 were designated by the Research Institute of Tuberculosis in Japan (Table 1). Each primer set was labelled with 4 kinds of fluorescent dye for capillary sequencer analysis. The capillary sequencer, a 3500 Genetic analyzer (Applied Biosystems, Foster City, CA, USA), was used for measuring the precise size of PCR products for each VNTR loci. However, in cases of large fragment sizes over 12,000 bp or an ambiguous size in the capillary sequencer, we measured the size by 1% agarose gel electrophoresis. Clusters were found as the result of comparison of genotyping data by BioNumerics version 5.1 software (Applied Maths). For PCR amplification, we mainly used Ex-Taq polymerase (Takara, Tokyo, Japan), except for Mix4, Mix7, and Mix9, which used KOD FX Taq polymerase (TOYOBO, Tokyo, Japan). PCR conditions were as follows: pre-denaturation for 5 minutes at 94℃, denaturation for 30 seconds at 94℃, annealing for 30 seconds at 63℃, extension for 1 minute at 72℃, and post-extension for 10 minutes at 72℃, except for Mix9 that had an annealing temperature of 60℃.
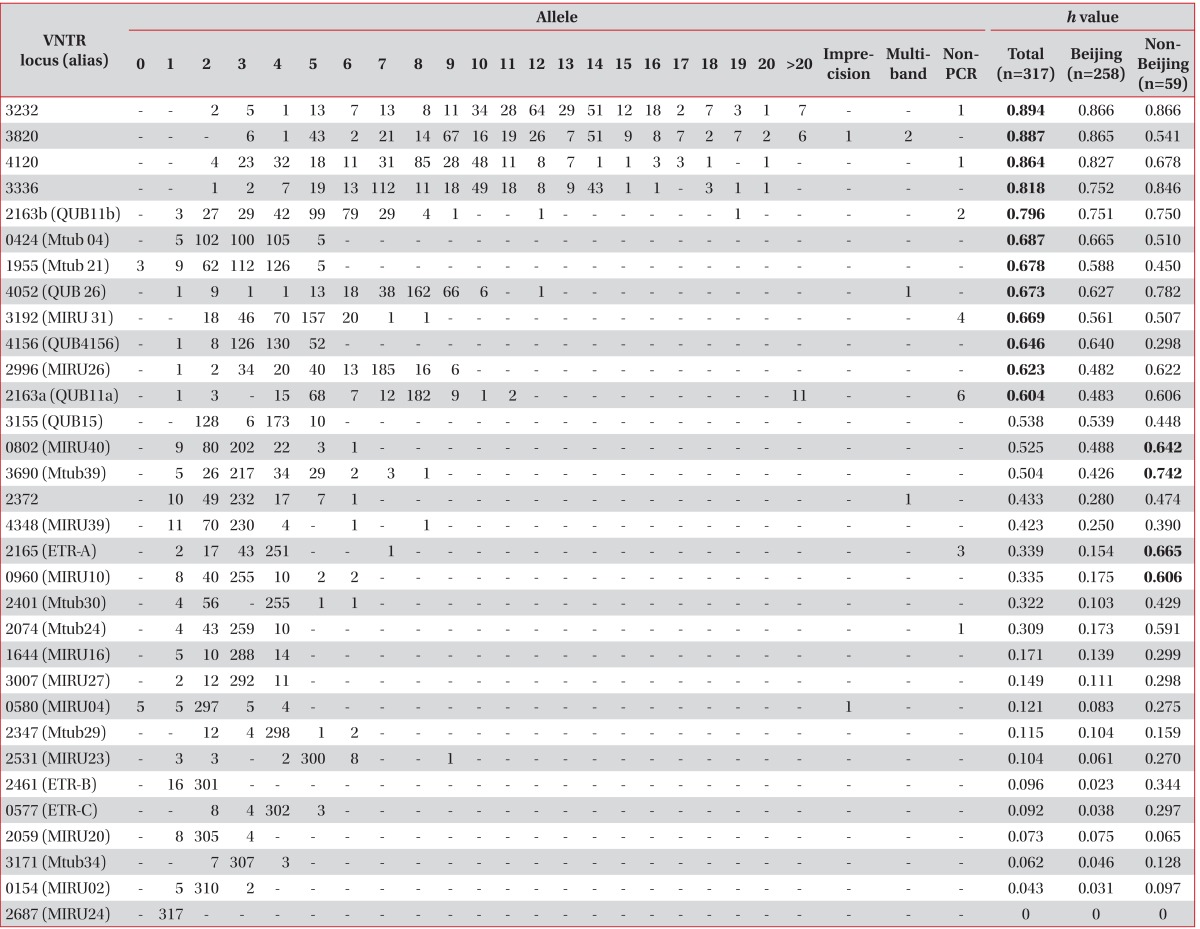
As a result of VNTR typing of the 317 M. tuberculosis strains isolated from Korea, VNTR loci showing a high h value of over 0.6 were VNTR 3232, 3820, 4120, 3336, 2163b, 0424, 1955, 4052, 3192, 4156, 2996, and 2163a. Interestingly, VNTR 0802, 3690, 2165, and 0960 revealed a high h value (over 0.6) only in non-Beijing M. tuberculosis isolates (Table 2).
Two PCR products amplified from VNTR 3802 and 0580 (MIRU04) were an unusual size, and were regarded as uncountable repeated numbers (Table 2). The imprecise PCR product of the VNTR 3802 loci was located between 8 and 9 copies, and that of VNTR 0580 was located between 3 and 4 copies of the repeated unit. Four PCR products in VNTR 3820, 4052 (QUB26), and 2372 had multiple bands. Non-amplified PCR products were found in VNTR 3232, 4120, and 2163b (QUB11b), 3192 (MIRU31), 2163a (QUB11a), 2165 (ETR-A), and 2074 (Mtub24).
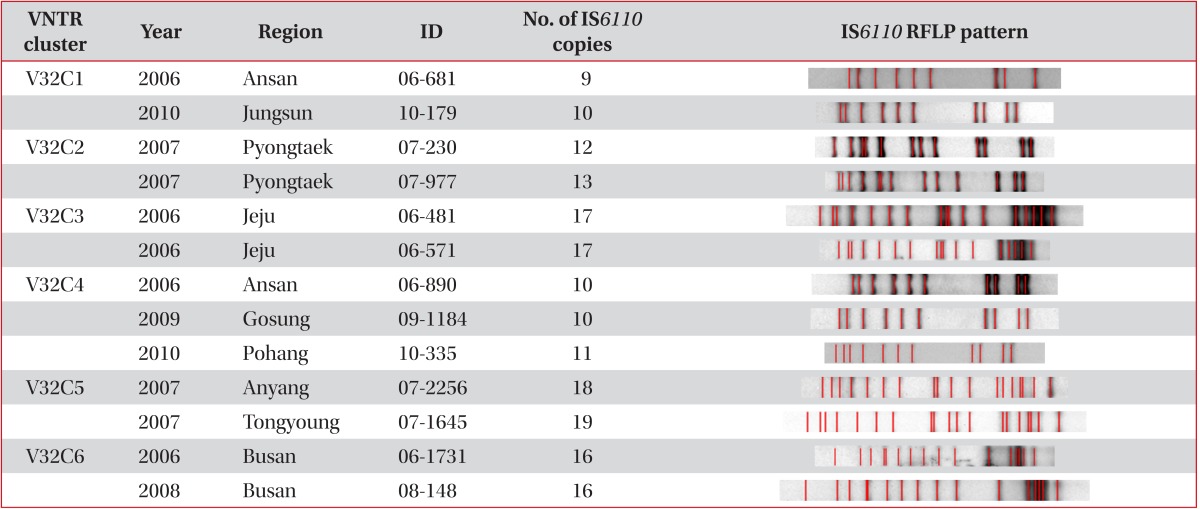
Six clusters in 13 patients were found after VNTR typing of the 32 loci, and only 3 clusters in 6 patients were found when additionally analysed by IS6110 RFLP typing (Table 3). Out of the 3 clusters, we found only one definite epidemiological linkage through personal contact. Patients with the V32C6 cluster were brother and sister. Patient 06-1731 with the cluster was the brother who developed TB in 2006, and was cured completely in 2007. Patient 08-148, who was the sister of patient 06-1731, had developed TB, and was cured in 2008. Even though random selection of PHC strains, we found the strains isolated from brother and sister TB-developed in different year by accident.
Two patients with V32C3 and IS6110 clusters had lived in Jeju province, even though they were not acquainted with each other. The two strains (06-890 and 09-1184) with the C32C4 cluster and identical IS6110 RFLP types were the K strain that is the most frequent endemic strain in Korea (4-5% in any TB population). The other strain (10-335) with the C32C4 cluster was the K family that exhibited a difference of only one band. Interestingly, another patient (10-179) with the V32C1 cluster also had the K strain. However, we found different copies in VNTR 2163a, 3232, and 3820 compared with that in V32C4 (data not shown).
When the internationally standardized 15 VNTR loci were applied to the 317 strains, we found 25 clusters in 83 strains (HGDI, 0.9958) (Table 4), and at least 7 VNTR loci (3232, 3336, 3820, 4120, 2372, 4348, and 3155) were needed to be equivalent to the discriminatory power of the 32 VNTR loci (data not shown). However, in cases of the hyper-variable 12 VNTR loci (3232, 3820, 4120, 3336, 2163b, 0424, 1955, 4052, 3192, 4156, 2996, and 2163a), 14 clusters in 32 strains (HGDI, 0.9995) were found, and we needed only 4 VNTR loci (4348, 2165, 0577, and 2372) to be equal to the discriminatory power of the 32 VNTR loci (data not shown). Furthermore, in the IS6110 RFLP typing, 25 clusters in 65 strains (HGDI, 0.9977) (Table 4) were obtained, and we needed at least 14 VNTR loci (4120, 3820, 2372, 1955, 0424, 4052, 3155, 2163b, 2163a, 0577, 0802, 4348, 3192, and 2996) for 3 clusters as a result of both the 32 VNTR loci and IS6110 RFLP typing (data not shown). When the 12 hyper-variable VNTR loci were applied to the 317 strains, followed by IS6110 RFLP typing, the discriminatory power was almost identical to the result of both the 32 VNTR loci and IS6110 RFLP typing (Table 4).
VNTR typing has been proposed as an alternative to IS6110 RFLP typing that bears intrinsic drawbacks for M. tuberculosis DNA typing. However, the disadvantage of VNTR typing is low discriminatory power with few combinations of VNTR loci. Therefore, we tried to find an optimal combination of DNA typing methods to discriminate M. tuberculosis isolated from Korea, which is accompanied by a high proportion of Beijing type3,4. Furthermore, compared with the surrounding countries, there is a higher proportion of RD181 among the ancient Beijing types4. These peculiar characteristics of Korean M. tuberculosis strains lead to an unclear distinction of M. tuberculosis isolates with only the 15 international standard VNTR loci.
Recently, the utility of hyper-variable VNTR loci has been used to compensate for the lack of discriminatory power of the 15 VNTR loci10,11,16. Hyper-variable VNTR loci, including 3232, 3820, 4120, 3336, and 2163a, also revealed a high h value in this study, which was similar to that in other studies9-11,16. In the report Murase et al.9, more than 4% of theses 5 loci had 15 or more copies, leading to difficulty in interpreting the exact copy number. We also found that 2-4% had 15 or more copies in VNTR 2163b, 3336, and 4120, and 13-16% had 15 or more copies in VNTR 3820, and 3232. These hyper-variable VNTR loci are excluded in the international standard 15 VNTR loci7 and JATA 129 because of the absence of PCR products, PCR products that are difficult to interpret with 15 or more copies, and amplification of multiple alleles. Non-amplification of PCR products not only occurred for hyper-variable VNTR loci but also general VNTR loci such as 2165 (ETR-A), and 2074 (Mtub24) in this study (Table 2). The major problem of hyper-variable VNTR loci was a high copy number of the repeats, which required additional analysis such as agarose gel electrophoresis. However, the copies in these hyper-variable VNTR loci are so diverse that they have a higher discriminatory power that is too valuable to exclude. Iwamoto et al.16 also recommended that these hyper-variable VNTR loci for second-line typing of clustering following the international standard 15 loci.
MIRU 40, Mtub 21, VNTR 4156, Mtub 04, QUB26, and QUB11b also revealed high h values in a previous study of South Korean M. tuberculosis19. In this study, we obtained a good result using hyper-variable VNTR loci, and IS6110 RFLP was useful as a secondary tool to discriminate the clusters.
An intriguing characteristic was found after comparing the mode of VNTR loci among Korea, Taiwan, and Japan9,20. Between Taiwan and Korea, there were some differences in the mode of copies of VNTR loci 0154, 3192, 2163b, 4052, 0424, 1955, 2347, and 2401. In particular, 5 copies of VNTR 0154, 0424, and 1955, and 3 copies of VNTR 2347 were modes in Taiwan but rare in Korea. Between Japan and Korea, there were some differences in VNTR 2163b, 1955, 3155, 3232, 3820, and 4120. Notably, 5 copies of VNTR 3155 and 4 copies of VNTR 3336 were more frequent in Japan but rare in Korea. These differences may be a clue for differentiation among the three countries. Compared with Beijing strains, we found that VNTR 2165, 0960, and 2074 loci had excellent higher h values for the non-Beijing strains, indicating that these VNTR loci may be useful to discriminate M. tuberculosis strains in countries with a high proportion of non-Beijing strains. The optimal combination of VNTR loci may be different depending on the proportion of Beijing strains or non-Beijing strains in each country.
We could not find any cluster consisted of only multi-drug resistant (MDR) strains in this study. The strains included in this study were selected randomly among strains collected from PHCs. Therefore, most of them (281 strains) were pansusceptible, only a few of strains (13 strains) were MDR, and the remains (23 strains) were any drug resistant with non-MDR.
Even though we analysed 32 VNTR loci for discrimination of 317 Korean M. tuberculosis isolates, we found 6 clusters. Among them, 3 clusters were not found to be clusters when additionally analysed by the IS6110 RFLP typing method, indicating that IS6110 RFLP typing is very useful for sub-classifying VNTR clusters. In terms of cost efficiency, it is difficult to use 32 VNTR loci for discrimination of M. tuberculosis, and the 15 international standard VNTR loci do not have satisfactory discrimination power for Korean strains. Inevitably, we need hyper-variable VNTR loci and the additional IS6110 RFLP typing method for effective discrimination of Korean M. tuberculosis strains.
The combination of 12 hyper-variable VNTR typing can be an effective tool for genotyping Korean M. tuberculosis isolates in which Beijing strains are predominant.
References
1. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993;31:406-409. PMID: 8381814.



2. Park YK, Bai GH, Kim SJ. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the western pacific region. J Clin Microbiol 2000;38:191-197. PMID: 10618086.



3. Park YK, Shin S, Ryu S, Cho SN, Koh WJ, Kwon OJ, et al. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J Microbiol Methods 2005;63:165-172. PMID: 15893392.


4. Kang HY, Wada T, Iwamoto T, Maeda S, Murase Y, Kato S, et al. Phylogeographical particularity of the Mycobacterium tuberculosis Beijing family in South Korea based on international comparison with surrounding countries. J Med Microbiol 2010;59(Pt 10):1191-1197. PMID: 20576748.


5. Cowan LS, Diem L, Monson T, Wand P, Temporado D, Oemig TV, et al. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol 2005;43:688-695. PMID: 15695665.



6. Smittipat N, Palittapongarnpim P. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber Lung Dis 2000;80:69-74. PMID: 10912281.


7. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006;44:4498-4510. PMID: 17005759.



8. Jiao WW, Mokrousov I, Sun GZ, Guo YJ, Vyazovaya A, Narvskaya O, et al. Evaluation of new variable-number tandem-repeat systems for typing Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J Clin Microbiol 2008;46:1045-1049. PMID: 18199785.



9. Murase Y, Mitarai S, Sugawara I, Kato S, Maeda S. Promising loci of variable numbers of tandem repeats for typing Beijing family Mycobacterium tuberculosis. J Med Microbiol 2008;57(Pt 7):873-880. PMID: 18566146.


10. Mokrousov I, Narvskaya O, Vyazovaya A, Millet J, Otten T, Vishnevsky B, et al. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J Clin Microbiol 2008;46:3576-3584. PMID: 18753356.



11. Mokrousov I, Vyazovaya A, Otten T, Zhuravlev V, Pavlova E, Tarashkevich L, et al. Mycobacterium tuberculosis population in northwestern Russia: an update from Russian-EU/Latvian border region. PLoS One 2012;7:e41318PMID: 22844457.



12. Iwamoto T, Grandjean L, Arikawa K, Nakanishi N, Caviedes L, Coronel J, et al. Genetic diversity and transmission characteristics of Beijing family strains of Mycobacterium tuberculosis in Peru. PLoS One 2012;7:e49651PMID: 23185395.



13. Kim SJ, Bai GH, Lee H, Kim HJ, Lew WJ, Park YK, et al. Transmission of Mycobacterium tuberculosis among high school students in Korea. Int J Tuberc Lung Dis 2001;5:824-830. PMID: 11573893.

14. Roring S, Scott A, Brittain D, Walker I, Hewinson G, Neill S, et al. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J Clin Microbiol 2002;40:2126-2133. PMID: 12037076.



15. Smittipat N, Billamas P, Palittapongarnpim M, Thong-On A, Temu MM, Thanakijcharoen P, et al. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J Clin Microbiol 2005;43:5034-5043. PMID: 16207958.



16. Iwamoto T, Yoshida S, Suzuki K, Tomita M, Fujiyama R, Tanaka N, et al. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol Lett 2007;270:67-74. PMID: 17302938.



17. Sun YJ, Bellamy R, Lee AS, Ng ST, Ravindran S, Wong SY, et al. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J Clin Microbiol 2004;42:1986-1993. PMID: 15131159.



18. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 1988;26:2465-2466. PMID: 3069867.



19. Shamputa IC, Lee J, Allix-Beguec C, Cho EJ, Lee JI, Rajan V, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol 2010;48:387-394. PMID: 20018816.


20. Chen YY, Chang JR, Huang WF, Kuo SC, Su IJ, Sun JR, et al. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on SNP and VNTR typing profiles in Asian countries. PLoS One 2012;7:e39792PMID: 22808061.






 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Print
Print Download Citation
Download Citation