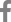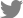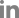|
 |
| Tuberc Respir Dis > Volume 79(2); 2016 > Article |
|
Abstract
Background
Lung cancer is the most common cause of cancer related death. Alterations in gene sequence, structure, and expression have an important role in the pathogenesis of lung cancer. Fusion genes and alternative splicing of cancer-related genes have the potential to be oncogenic. In the current study, we performed RNA-sequencing (RNA-seq) to investigate potential fusion genes and alternative splicing in non-small cell lung cancer.
Methods
RNA was isolated from lung tissues obtained from 86 subjects with lung cancer. The RNA samples from lung cancer and normal tissues were processed with RNA-seq using the HiSeq 2000 system. Fusion genes were evaluated using Defuse and ChimeraScan. Candidate fusion transcripts were validated by Sanger sequencing. Alternative splicing was analyzed using multivariate analysis of transcript sequencing and validated using quantitative real time polymerase chain reaction.
Results
RNA-seq data identified oncogenic fusion genes EML4-ALK and SLC34A2-ROS1 in three of 86 normal-cancer paired samples. Nine distinct fusion transcripts were selected using DeFuse and ChimeraScan; of which, four fusion transcripts were validated by Sanger sequencing. In 33 squamous cell carcinoma, 29 tumor specific skipped exon events and six mutually exclusive exon events were identified. ITGB4 and PYCR1 were top genes that showed significant tumor specific splice variants.
Lung cancer is the most common cause of cancer related death. Alterations of sequence or structure of genes and their expression have an important role in the pathogenesis of lung cancer. Fusion genes and alternative splicing of cancer-related genes have the potential to gain oncogenic activity.
Fusion genes can potentially be used for lung cancer diagnosis, prognosis, and therapy. EML4-ALK fusion gene gains oncogenic activity by fusing two genes, one that has a role as a dimerization factor and other as a tyrosine kinase, and the oncogenic activity can be prevented by a kinase inhibitor1. Recent advances in sequencing technology enabled analysis of genetic changes, and there already has been several data reported related to lung cancer using the sequencing technology2,3.
The recent developments of next-generation sequencing allow for increased base coverage of a DNA sequence, as well as higher sample throughput. This has facilitated the reconstruction of the entire transcriptome by deep RNA sequencing (RNA-seq), even without a reference genome4. It provides the ability to look at alternative gene spliced transcripts, posttranscriptional modifications, gene fusion, mutations/single-nucleotide polymorphism, and changes in gene expression.
Alternative splicing of cancer-related genes can affect cell cycle control, signal transduction pathway, apoptosis, angiogenesis, invasion, and metastasis5. Five different types of alternative splicing affect the resulting translated protein products6. Recent advance in RNA-seq provides the opportunity to quantitatively study alternative splicing7. Splice isoform can also be a therapeutic target8.
In the current study, we performed RNA-seq to investigate potential oncogenic alternative splicing and fusion genes in 86 pairs of tissue samples from non-small cell lung cancer and normal lung.
This study included tissues obtained from the Biobank of Asan Medical Center (Seoul, Korea) donated by 88 male smokers who underwent surgery for non-small cell lung carcinoma (NSCLC) between March 2008 and March 2011. All of the paired NSCLC and adjacent normal tissue specimens used in this study were acquired from surgical specimens. Cancer and normal tissue specimens were grossly dissected and preserved in liquid nitrogen immediately after surgery. All protocols were approved by the Institutional Review Board of Asan Medical Center (2011-0711) and Kangwon National University Hospital (2011-04-004).
Resected tumor specimens were evaluated by routine frozen section procedures. The study samples were snap-frozen and stored at -80℃. Tumor and normal lung tissues were selected by a pathologist using manual microdissection under an inverted microscope.
For RNA-Seq, we extracted RNA from tissue using an RNeasy 96 Universal Tissue Kit (Qiagen, Gaithersburg, MD, USA). Total RNA quality and quantity were verified spectrophotometrically (NanoDrop 1000 Spectrophotometer; Thermo Scientific, Wilmington, DE, USA) and electrophoretically (Bioanalyzer 2100; Agilent Technologies, Palo Alto, CA, USA). To construct Illumina-compatible libraries, a TruSeq RNA Library Preparation Kit (Illumina, San Diego, CA, USA) was used according to the manufacturer's instructions. In brief, messenger RNA purified from total RNA using polyA selection was chemically fragmented and converted into single-stranded cDNA using random hexamer priming. Double-stranded (ds) cDNA was generated for TruSeq library construction. Short ds-cDNA fragments were joined with sequencing adapters, and suitable fragments were separated by agarose gel electrophoresis. TruSeq RNA libraries constructed by polymerase chain reaction (PCR) amplification were quantified using quantitative PCR (qPCR) according to the qPCR Quantification Protocol Guide, and their quality was assessed electrophoretically (Bioanalyzer 2100; Agilent Technologies). Sequencing was performed using a HiSeq 2000 platform (Illumina).
To discover gene fusion from RNA-seq data, we used DeFuse version 0.4.3 and ChimeraScan version 0.4.59,10.
In order to validate fusion transcript by Sanger sequencing, fusion candidate were selected. Fusion transcripts were observed only in cancer tissues, and protein coding transcripts were selected. Genes that were reported in cancer gene database (COSMIC, ChimerDB 2.0) and previous studied were validated.
For Sanger sequencing, 2 µg of total RNA was used for cDNA synthesis with an oligo-dT primer and PrimeScript reverse transcription polymerase chain reaction Kit (Takara, Kyoto, Japan) according to the manufacturer's protocol.
Fusion gene specific primer pairs and TAKARA Ex-Taq polymerase (Takara) were used for the PCR reaction. After purification, PCR products were sequenced with the BigDye Terminator v3.1 Sequencing Kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA, USA). All DNA sequenced comparison alignments were performed using DNAstar SeqMan program (DNAstar, Madison, WI, USA).
To estimate alternative spliced transcripts, the RNA-seq reads were mapped to the human genome using TopHat version 1.3.311. Alternative splicing events were detected using MATS 3.0.612. The statistical model calculated the p-value and false discovery rate by the Benjamini-Hochberg method that the difference in the isoform ratio of a gene between two conditions.
Demographic characteristics of subjects are listed (Table 1). A total of 86 subjects participated; all were male. Fifty-three were diagnosed with adenocarcinoma and 33 with squamous cell carcinoma (SqCC). The average age of subject was 61.1±9.4 and the average of smoking pack-year was 34.5±17.5. All analysis was processed in normal-cancer paired tissue samples.
In the fusion gene analysis, 86 samples were analyzed using DeFuse and 33 SqCC samples were analyzed using both DeFuse and ChimeraScan. To identify expressed fusion-genes, we used DeFuse and ChimeraScan. From the RNA-seq data, 1,293 and 6,455 fusion transcripts were detected by the two different programs, respectively. Two EML4-ALKs and one SLC34A2-ROS1 fusion gene, already known to be oncogenic, were detected in analysis result of DeFuse1,2.
From these results, one EML4-ALK and one SLC34A2-ROS1 fusion genes were validated by Sanger sequencing. Also according to our procedure, four fusion transcripts were selected (Table 2). The frequencies of the selected fusion transcripts were detected in 1%-5% of all samples. The four fusion transcripts were validated by Sanger sequencing (Figure 1).
In the alternative splicing analysis, 33 SqCC samples were analyzed among the 86 samples. To identify alternative splicing events, total reads and reads aligned of alternative splicing were determined (Table 3).
There were 37 differential skipped exon events and six mutually exclusive exon events in the cancer samples compared to the normal samples. Also from these results, we found CD44 and vascular endothelial growth factor A, which were already known as alternative spliced genes, to exist in the cancer samples, and the most significant gene was CD44 (Table 4)5,13. As a result of comparing the normal-cancer individual paired samples, there were 12,069 differential skipped exon events. To obtain selective list, 12,069 events were filtered with a condition (normal sample SE count, 0; tumor sample SE count, ≥10; and SE covering sample, ≥30), then 29 differential skipped exon events were selected. In the list of the selected genes, there were ITBG4 and PYCR1 outstanding genes and the most significant gene is ITGB4 (Table 4).
In the current study, we have identified candidate fusion genes and alternative splicing in non-small cell lung cancer.
In the present study, EML4-ALK was detected with DeFuse, whereas it was not detected with ChimeraScan. DeFuse is more focused on finding breakposition of fusion candidates and applies various statistical methods and database to filter out fusion candidates, while ChimeraScan concentrates more on finding genes of fusion candidates. Therefore, transcripts from DeFuse were mainly used and those from ChimeraScan were used complementally. One EML4-ALK fusion gene and one SLC34A2-ROS1 fusion gene were detected only in our cancer samples and not detected in the normal samples.
Potential candidate fusion transcripts identified in the present study are AL137145.2-PFKFB3, C4orf3-KLHL2, TPPPBRD9, and HNRNPA2B1-SKAP2. AL137145.2-PFKFB3 fusion was detected with the highest percentage in four samples among the 86 cancer samples (4.7%). According to NCBI, SMART and Ensemble database, AL137145.2 was discovered simply as a protein coding gene and has not been identified clearly yet. 6-Phosphofructo-2-kinase (PFKFB3) has a PGAM domain which is involved with the transfer of phosphorous groups. There are no reports on fusion of PFKFB3. However, there has been many previous studies that show tumor growth is suppressed by the inhibition of PFKFB3 activity14,15. Therefore, it is possible to formulate the hypothesis that changes in PFKFB3 by fusing with AL137145.2 could be involved with cancer. C4orf3-KLHL2 fusion was found in two samples out of the 86 cancer samples. Kelch-like protein 2 (KLHL2) consists of BTB/POS, BACK, and repeated Kelch domains. These domains were involved in evolution and ubiquitination, and they are known to be associated with cancer16. C4orf3-KLHL2 fusion transcript could possess oncogenic activity by changes in the KLHL2 domain. TPPP-BRD9 and HNRNPA2B1-SKAP2 were detected in 1.2% of the total samples (1 out of 86), and the two fusions were found by both programs. In the case of TPPP-BRD9, there are no outstanding studies about tubulin polymerization promoting protein (TPPP) and bromodomain-containing protein 9 (BRD9) fusion. However, bromodomain family is used as a target of cancer therapy. BRD4 was reported to have a role in breast cancer progression and metastasis17. TPPP-BRD9 fusion transcript was constructed by combining exon1 of BRD9 with exon 11 of TPPP .
Heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) of the HNRNPA2B1-SKAP2 fusion transcript was found to fuse with several other partner genes such as ETV1, AUTS2, PRR13, and SMARCA2 to form cancer-related fusion genes in COSMIC and ChimerDB2.018. Similar to these reports, our results from RNA-seq showed HNRNPA2B1 fused with different gene partners, such as src kinase associated phosphoprotein 2 (SKAP2), nuclear factor, erythroid 2-like 3 (NFE2L3), and pterin-4-alpha-carbinolamine dehydratase 2 (PCBD2). SKAP2 was reported to negatively regulate cell migration and tumor invasion in fibroblasts and glioblastoma cells19. HNRNPA2B1-SKAP2 was organized by fusing exon 5 of SKAP2 next to exon 8 of HNRNPA2B1. This configuration is the form that included all key domains as RNA recognition motif and sarcoma homology 3 domain. Therefore, HNRNPA2B1-SKAP2 may have the potential to be a fusion gene of cancer.
In the alternative splicing of this study, exon 6 or 12 skipped CD44 and exon 6 or 7 skipped vascular endothelial growth factor (VEGF), which are already known splicing variants, were identified. Many previous studies have investigated the roles of various CD44 isoforms associated with cancer. Banky et al.20 reported the roles of CD44 according to alternative splice patterns that were detected not only in colorectal tumor but also in other tumor tissues. CD44 isoforms, containing variable protein domain v3 and v6, have a role in metastasis and affects metastatic development. Correlation exists between tumor progression and higher expression levels of variable protein domain v3 and v6 in quantitative assessment20. VEGF is a regulator of angiogenesis and is strongly involved in cancer. Zygalaki et al.21 studied previously about the correlation between its expression pattern and clinic-pathological characteristics of tumors. Exon 6 or 7 skipped VEGFA splice variants were detected in our analysis result, which is the same as prior reports.
This study discovered novel splice variant forms of ITGB4 (integrin, beta 4) and pyrroline 5-carboxylate reductase 1 (PYCR1). ITGB4 is known to be over-expressed in tumor cells and has a role in metastasis22. Also, ITGB4 was found to skip exon 35 in about half of patient samples23. Exon 18, 19, 33, or 34 skipped ITGB4 splice variants were found in our samples. Skipping of exon 18, 19, and 34 were novel variant forms. Jariwala et al.24 reported that PYCR1 is one of the novel androgen receptor target genes in prostate cancer. However, alternative splicing of PYCR1 was not reported in lung cancer. PYRC1 exon 2 or 3 skipped PYCR1 splice variants were detected in this study, and the exon 2 skipped form was a novel variant form.
A limitation of the current study is a lack of a functional study. Oncogenic potential of fusion transcripts and the role of alternative splicing should be investigated in the future.
In conclusion, novel potential fusion transcripts and splice variants were identified in NSCLC. Their functional significance in the pathogenesis of lung cancer should be evaluated.
Acknowledgements
This study was supported by a grant from the National R&D Program for Cancer Control (1020420) and from the National Project for Personalized Genome Medicine (A111218-11-GM02), Ministry for Health and Welfare, Republic of Korea.
This work was supported by 2015 Research Grant from Kangwon National University (520150333).
References
1. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-566. PMID: 17625570.



2. Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-2119. PMID: 22975805.



3. Kim SC, Jung Y, Park J, Cho S, Seo C, Kim J, et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS One 2013;8:e55596PMID: 23405175.



4. Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet 2011;12:671-682. PMID: 21897427.



5. Kim YJ, Kim HS. Alternative splicing and its impact as a cancer diagnostic marker. Genomics Inform 2012;10:74-80. PMID: 23105933.



6. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003;72:291-336. PMID: 12626338.


7. Feng H, Qin Z, Zhang X. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett 2013;340:179-191. PMID: 23196057.


8. Miura K, Fujibuchi W, Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis 2012;33:2311-2319. PMID: 23118106.



9. McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 2011;7:e1001138PMID: 21625565.



10. Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011;27:2903-2904. PMID: 21840877.




11. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-1111. PMID: 19289445.




12. Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 2012;40:e61PMID: 22266656.




13. Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu N, McGlade CJ, Tsao MS, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol 2011;31:138-150. PMID: 21041478.


14. Bando H, Atsumi T, Nishio T, Niwa H, Mishima S, Shimizu C, et al. Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin Cancer Res 2005;11:5784-5792. PMID: 16115917.


15. Clem BF, O'Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA 2nd, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther 2013;12:1461-1470. PMID: 23674815.



16. Dhanoa BS, Cogliati T, Satish AG, Bruford EA, Friedman JS. Update on the Kelch-like (KLHL) gene family. Hum Genomics 2013;7:13PMID: 23676014.



17. Alsarraj J, Hunter KW. Bromodomain-containing protein 4: a dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix. Int J Breast Cancer 2012;2012:670632PMID: 22295248.



18. Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakacs A, Coppola L, et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res 2011;71:4464-4472. PMID: 21586613.


19. Shimamura S, Sasaki K, Tanaka M. The Src substrate SKAP2 regulates actin assembly by interacting with WAVE2 and cortactin proteins. J Biol Chem 2013;288:1171-1183. PMID: 23161539.


20. Banky B, Raso-Barnett L, Barbai T, Timar J, Becsagh P, Raso E. Characteristics of CD44 alternative splice pattern in the course of human colorectal adenocarcinoma progression. Mol Cancer 2012;11:83PMID: 23151220.



21. Zygalaki E, Tsaroucha EG, Kaklamanis L, Lianidou ES. Quantitative real-time reverse transcription PCR study of the expression of vascular endothelial growth factor (VEGF) splice variants and VEGF receptors (VEGFR-1 and VEGFR-2) in non small cell lung cancer. Clin Chem 2007;53:1433-1439. PMID: 17599955.



22. Giancotti FG. Targeting integrin beta4 for cancer and antiangiogenic therapy. Trends Pharmacol Sci 2007;28:506-511. PMID: 17822782.


23. Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics 2006;7:325PMID: 17192196.



24. Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, Cogan JP, et al. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer 2007;6:39PMID: 17553165.



Figure 1
Confirmation by Sanger sequencing of fusion transcript structure according to presence of exon of genes. (A) PFKFB3-AL137145.2. (B) KLHL2-C4orf3. (C) TPPP-BRD9. (D) HNRNPA2B1-SKAP2. The black arrows indicate orientations.
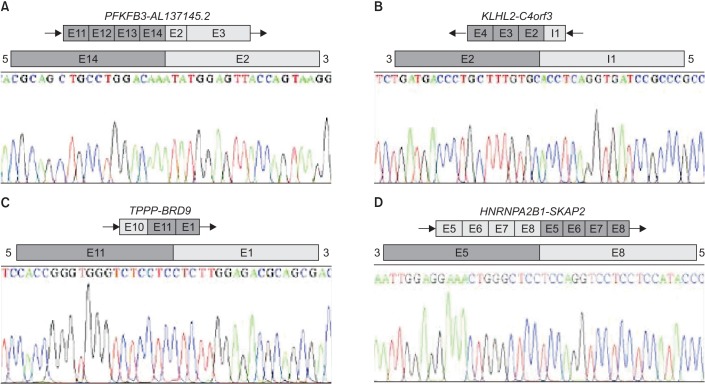
Table 1
Baseline characteristics of the subjects
Table 2
The list of fusion transcripts from RNA sequencing and validation results of fusion by Sanger sequencing
Table 3
Statistics based on per group analysis using Burrows-Wheeler Alignment for paired-end reads
| Sample | Total alignment | |||
|---|---|---|---|---|
| Total reads | Reads aligned | Reads aligned in pairs | Reads AlignedTo DiffChr (mapQ≥5) | |
| Normal | 50,590,052 | 42,607,206 | 37,569,185 | 258,633 |
| Tumor | 49,743,632 | 42,318,389 | 37,636,657 | 299,863 |


 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Print
Print Download Citation
Download Citation