|
 |
| Tuberc Respir Dis > Volume 86(1); 2023 > Article |
|
Abstract
Background
There is a global increase in isolation of nontuberculous mycobacteria (NTM). The aim of the study was to analyze longitudinal trends of NTM identification and pattern of antimicrobial susceptibility testing.
Methods
NTM recovery rates, distribution of NTM species identification, and antimicrobial susceptibility pattern of NTM at Pusan National University Yangsan Hospital between January 2016 and December 2020 were retrospectively analyzed.
Results
A total of 52,456 specimens from 21,264 patients were submitted for mycobacterial culture, of which 2,521 from 1,410 patients were NTM positive over five years (January 2016 to December 2020). NTM isolation showed an increasing trend from 2016 to 2020 (p<0.001, test for trend) mainly caused by Mycobacterium avium complex. The vast majority of M. avium complex were susceptible to key agents clarithromycin and amikacin. For Mycobacterium kansasii, resistance to rifampin and clarithromycin is rare. Amikacin was the most effective drug against Mycobacterium abscessus subspecies abscessus and Mycobacterium subspecies massiliense. Most of M. subspecies massiliense were susceptible to clarithromycin, while the majority of M. abscessus subspecies abscessus were resistant to clarithromycin (p<0.001).
Nontuberculous mycobacteria (NTM) are defined as a group of Mycobacterium species other than Mycobacterium tuberculosis and Mycobacterium leprae [1]. Unlike M. tuberculosis that causes tuberculosis (TB) or M. leprae that causes leprosy, NTM are opportunistic pathogens that are widespread in the environments such as soil and water [1]. An increase in the number of NTM isolation has been observed globally [2-6]. This is partly explained not only by increased awareness of symptoms caused by NTM infection and enhancements in diagnostic techniques, but also by increased numbers of susceptible hosts and the ubiquitous presence of NTM [7]. The situation is expected to become worse because most NTM either inherently possess or have the ability to acquire resistance against conventional antibiotics [8].
Currently, macrolide-based antibiotics such as clarithromycin and azithromycin are the first choice of drugs for treating most NTM infections [9]. However, treatment regimens vary by species. The regime also includes ethambutol and rifampin for slowly growing NTM [10], while it adds an aminoglycoside and either cefoxitin, imipenem, or tigecycline for rapidly growing NTM [11]. Treatment duration is prolonged up to 18 months [12]. A complete cure is not always possible [4,13]. The role of antimicrobial susceptibility testing (AST) in guiding treatment remains a subject of debate [14]. Discordance between in vitro AST and in vivo drug activity has been observed for some drugs and NTM species [12,15]. For examples, although ethambutol, rifampin, and rifabutin are clinically useful for Mycobacterium avium complex (MAC), the relationship between in vitro susceptibility results and in vivo response to antibiotics is controversial and poorly understood [16,17]. Two contradictory studies have been conducted in Korea on the relationship between in vitro AST results and treatment outcomes of ethambutol and rifampin [18,19]. AST of the second line drugs moxifloxacin and linezolid might also be taken into consideration for macrolide-resistant MAC isolates and/or isolates from patients who cannot tolerate macrolide therapy using breakpoints in Clinical and Laboratory Standards Institute (CLSI) document M62 [16]. However, the in vivo usefulness of these drugs remains unknown [20]. In addition, the evidence is limited regarding the use of in vitro AST-based treatment with injectable amikacin and beta-lactams for patients with Mycobacterium abscessus subspecies abscessus infection [21]. For Mycobacterium kansasii, there is no correlation between in vivo AST and in vivo clinical outcomes for any drugs other than rifampin [22]. These discrepancies partly originate from laboratory technical difficulties associated with AST, standardization of methods, and a lack of clinical validation [14]. Nevertheless, laboratory tests for determining AST of NTM can confirm the initial drug treatment choice and any emerging drug resistance [20]. In addition, AST may provide additional information for different drug treatment choices [20]. The prevalence of intrinsic and acquired drug resistance can also be estimated based on NTM AST in a community [20]. The goal of this study was to analyze patterns of AST for NTM isolated in a Korea tertiary university hospital and longitudinal trends of NTM identification.
Mycobacterial culture and AST results were obtained from the electric medical records system from the clinical microbiology laboratory of Pusan National University Yangsan Hospital. Data were obtained for all specimens received between January 2016 and December 2020. These specimens were either from patients with presumptive mycobacterial infection or patients with confirmed TB or NTM infection. Various clinical specimens including pulmonary and extrapulmonary samples were included in this study. This study was approved by Pusan National University Yangsan Hospital Institutional Review Board (05-2021-237). Written informed consent by the patients was waived due to a retrospective nature of our study.
Clinical specimens were prepared according to a standard protocol [23]. Processed specimens were inoculated into BACTEC 960 mycobacterial growth indicator tubes (MGIT) (Becton Dickinson, Franklin Lakes, NJ, USA) and Ogawa media (Eiken, Tokyo, Japan) for mycobacterial culture. Liquid cultures were kept for 6 weeks. Solid cultures were examined for 8 weeks. Positive acid-fast bacilli (AFB) cultures were confirmed using Ziehl-Neelsen staining. TB/NTM differentiation was performed with an MPT 64 Ag test (SD Bioline Kit, Standard Diagnostics, Yongin, Korea) and an AdvanSure TB/NTM real-time PCR kit (LG Chem, Seoul, Korea).
Positive NTM isolates were sent to Korean Institute of Tuberculosis (KIT) for species identification and AST. NTM species identification was performed using an AdvanSure Mycobacteria GenoBlot assay (LG Chem) at KIT. The assay can identify M. tuberculosis complex and 20 different NTM species [24]. The reverse line probe assay can detect the presence of ≥2 NTM species infection in a single specimen. NTM species that could not be differentiated with the assay were confirmed by multigene sequence-based typing. Sequencing results of 16s rRNA, rpoB, and hsp65 were analyzed according to the CLSI guideline MM18-ED2 [25]. Unclassified NTM were given to NTM species that failed to be differentiated at species levels after sequencing. Discrimination between M. abscessus subspecies abscessus and M. abscessus subspecies massiliense was performed using an ERM-plus real-time PCR kit (LG Chem; commercially not available). The kit not only can differentiate between M. abscessus subspecies abscessus and M. abscessus subspecies massiliense, but also identify infection of both species in a single specimen. Mixed infection was defined as simultaneous identification of ≥2 species in a single specimen.
At KIT, AST was performed by the reference broth microdilution method for slowly and rapidly growing NTM against different drugs according to the CLSI guideline [20]. Minimum inhibitory concentration (MIC) values were analyzed using Muller-Hinton broth in polystyrene 96-well plates containing drugs in 2-fold increasing concentrations (μg/mL) (Supplementary Table S1). MIC values were categorized as susceptible (S), intermediate (I), or resistant (R) in accordance with the CLSI guideline [16] (Supplementary Table S1). To evaluate inducible resistance to clarithromycin for M. abscessus subspecies abscessus and M. abscessus subspecies massiliense, MIC values were established both at days 3 and 14 in accordance with the CLSI guideline [20]. Clinical specimens that had mixed NTM infections were also subjected to AST at KIT. However, obtaining pure culture isolates of single species was not performed prior to the broth microdilution test. We believed that this AST method for specimens was inappropriate. Hence, AST results from clinical specimens with mixed NTM infection were excluded from the current study.
The number of specimens positive for AFB culture and the proportion of NTM isolates recovered from positive specimens were calculated for each year. A linear-by-linear association exact test was used to test for significant trends. Chi-squared test and Fischer’s exacttest were used to compare differences in antimicrobial susceptibility to each drug between NTM species using SPSS for Windows version 26.0 (IBM, Armonk, NY, USA).
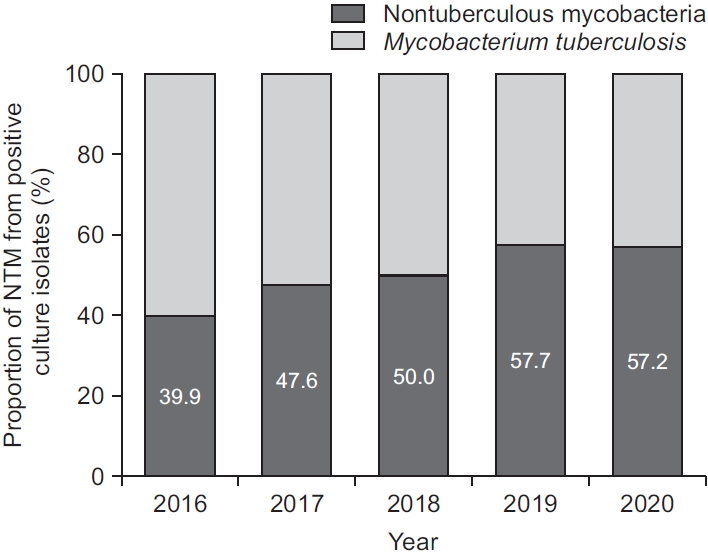
Data were obtained for 52,456 clinical specimens from 21,264 patients during the study period. The number of specimens received for mycobacterial culture steadily increased from 9,435 in 2016 to 11,396 in 2019, but decreased to 10,309 in 2020 (Figure 1). While the number of specimens positive for culture for M. tuberculosis was continuously decreased from 594 (6.3% of total specimens, 60.1% of all positive culture) in 2016 to 381 (3.7% of total specimens, 42.8% of all positive culture) in 2020, the number of NTM isolates was gradually increased from 395 (4.2% of total specimens) in 2016 to 590 (5.2% of total specimens) in 2019 (Figure 1). In total, 5,005 (9.5%) from 2,356 patients were positive for mycobacterial culture. Of these 5,005 culture positive specimens, 2,521 from 1,410 patients were positive for NTM. The proportion of NTM recovered from specimens showed an increasing trend from 39.9% in 2016 to 57.2% in 2020 (p<0.001, test for trend). Interestingly, culture positivity and proportions of NTM recovered from culture isolates were both decreased from 2019 to 2020 (Figures 1, 2).
Of the 2,521 NTM cultures, species identification was obtained for 718 specimens (28.5% of 2,521) from 646 patients. Mycobacterium intracellulare (45.8%) was the most commonly isolated NTM, followed by M. avium (21.4%) and M. abscessus subspecies abscessus (7.9%) (Table 1). Interestingly, 8.5% of identified clinical isolated showed mixed NTM infection, of which a combination of M. intracellulare and M. abscessus subspecies abscessus (1.8%) was the most common, followed by mixed infection of M. intracellulare and M. avium (1.4%) (Table 1). Annual trends in distribution of NTM species recovered from clinical isolates are shown in Figure 3. The proportion of every NTM species was comparable by year (p≥0.05).
AST was obtained for 387 clinical specimens from 352 patients. Every isolate of M. intracellulare was susceptible to clarithromycin. Only one of 91 isolates of M. avium was resistant to clarithromycin (Table 2). It was found that 88% (191/217) of M. intracellulare isolates and 76% (69/91) (p=0.01) of M. avium isolates showed amikacin susceptibility. There was a statistically significant difference in the extent of susceptibility to moxifloxacin and linezolid between M. intracellulare and M. avium (Table 2).
For M. kansasii, 96% (22/23) of isolates showed rifampin susceptibility, while 100% (23/23) exhibited susceptibility to clarithromycin (Table 3). In general, M. kansasii showed high rates of susceptibility towards most test drugs except for ciprofloxacin and doxycycline.
Amikacin was the most effective drug against M. abscessus subspecies abscessus (92%, 35/38) and M. abscessus subspecies massiliense (100%, 18/18) (Table 4). It was found that 26% (10/38) of M. abscessus subspecies abscessus were susceptible to clarithromycin at both day 3 and day 14. At day 3, only 3% (1/38) of M. abscessus subspecies abscessus exhibited clarithromycin resistance, which was indicative of acquired resistance to the drug. It was found that 71% (27/38) of M. abscessus subspecies abscessus were susceptible or intermediate at day 3 but resistant to clarithromycin at day 14, which correlated with inducible resistance to clarithromycin. Conversely, none of M. abscessus subspecies massiliense isolates exhibited inducible resistance to clarithromycin. Instead, they all displayed the same susceptibility results at days 3 and 14.
In accordance with past studies both from Korea and other countries [2-5], our study showed that the number of NTM isolates had increased over time. Currently, NTM constitute the majority of mycobacterial culture isolates in our hospital. Reasons behind this increase of NTM isolation are yet to be fully elucidated. Possible theories include awareness of NTM as pathogens, advancements of laboratory detection methods, increased environmental exposures, and increased numbers of susceptible patients with predisposing factors [26]. Interestingly, the number of specimens for mycobacterial culture, the number of NTM isolates, and the proportion of NTM were slightly decreased from the previous year to 2020 (Figure 1). A recent study conducted in Korea has shown that environmental exposure such as public bath is a risk factor for NTM disease [27]. The ongoing pandemic of coronavirus disease 2019 (COVID-19) has drastically changed the hygiene and sanitation procedures in public places in Korea, which might have affected patients in a way that they were less exposed to ubiquitous presence of NTM. In addition, since the number of specimens for mycobacterial culture was decreased from 2019 to 2020 (Figure 1), some patients might be less eager to seek a medical service during the pandemic. COVID-19-related social restrictions intended to control the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) may also have been associated with decreased rates of other infectious diseases such as TB and HIV [28,29]. Our findings suggest that COVID-19 might affect recovery rates of NTM in a clinical microbiology laboratory of a tertiary hospital.
In our study, M. intracellulare and M. avium were the most common organisms recovered from clinical specimens followed by M. abscessus subspecies abscessus, consistent with previous reports from Korea [3,30]. Geographic distribution of NTM species recovered from clinical isolates can vary. For example, MAC was the most common NTM species isolated in the United States [31], whereas M. kansasii and M. abscessus complex (MABC) were the most frequent species isolated in Singapore [32] and Shanghai China [33], respectively. Such differences in the distribution of NTM species around the world can be in part due to variations in sample sources, climate changes, and population density [34]. In this study, 8.5% of all clinical specimens contained mixed infection. This proportion was similar to 7.5% in a study conducted in southwest China [35], but different from 30.1% in a study performed in Singapore [36]. Studies on isolation rates of mixed NTM infection in Korea are lacking. These results indicate that distribution of NTM multispecies infection could vary geographically as single NTM species. Although the clinical significance of mixed NTM species infection from the same patient is not fully understood yet, it is a frequent finding of NTM lung infection [35]. Disease status and clinical outcomes of patients with mixed NTM infection might be different from those with a single NTM infection [37]. Unfortunately, the correlation between mixed NTM infection and patient outcomes could not be investigated in our study due to the lack of data pertaining to clinical treatment. Misidentification of mixed NTM infection is not an uncommon finding, which can cause a critical problem in patient management [37]. Therefore, future studies should elucidate clinical outcomes of mixed NTM infection with accurate diagnosis of single or mixed NTM infection.
It has been reported that a vast majority of NTM show resistance against anti-mycobacterial drugs [38]. Although the association between in vitro AST patterns of NTM and in vivo patient outcomes was poor in previous studies [39,40], the evidence of clinical impacts of AST was observed for rifampin in M. kansasii infection [41], macrolides and amikacin in MAC infection [42,43], and macrolides in MABC [44]. CLSI’s recommendations for selecting antimicrobial agents are based on published studies about appropriate therapy [20]. Although the exact role of AST for NTM remains to be elucidated by larger clinical studies, clinical benefits of AST for NTM are manifolds [20]. First, AST results can guide treatment choices for NTM infection. Second, AST profiles might lead to better prediction of treatment outcomes. Third, detailed studies of NTM AST can be accessed by a community to estimate the prevalence of drug resistance.
MAC primary consists of M. intracellulare and M. avium. Clarithromycin and amikacin are the two first line drugs against MAC [20]. In our study, most of MAC isolates showed susceptibility to clarithromycin and amikacin, consistent with their roles in most NTM treatment regimens and correlation between in vitro susceptibility and in vivo clinical outcomes [4,42,43]. Although moxifloxacin is recommended by the CLSI as the second line drug of AST for MAC, clinical evidence is limited for clinical efficacy and safety of moxifloxacin for MAC pulmonary disease [44]. In addition, no correlation was observed between in vitro and in vivo response to moxifloxacin [45]. Clinical efficacy of linezolid, another second drug for MAC, is also uncertain [46]. According to our study, both drugs had limited activity against MAC isolates, as described in other studies [47-50].
A multidrug antibiotic regimen comprising of rifampin, ethambutol, and isoniazid is currently the recommended course of treatment for M. kansasii [46]. MIC values and clinical outcomes for ethambutol and isoniazid do not correlate well. Therefore, the current CLSI guideline does not recommend reporting in vitro AST for the drugs [16]. In addition, a short-course therapy that substitutes a macrolide for isoniazid has been proposed. However, its effectiveness has only been examined in limited studies [51]. In our study, most of M. kansasii isolates were susceptible to tested drugs except for ciprofloxacin and doxycycline, in agreement with another study [52].
MABC includes three subspecies, M. abscessus subspecies abscessus, M. abscessus subspecies bolletti, and M. abscessus subspecies massiliense [53]. Since some drugs’ susceptibility testing outcomes can only be applied to particular species, the CLSI and the majority of mycobacterial experts strongly suggest species-level identification or, in the case of the MABC, subspecies-level identification [46]. This is also required for the appropriate selection of treatment choices [46]. Our study showed that the vast majority of MABC isolates were susceptible to amikacin. For clarithromycin, we demonstrated that M. abscessus subspecies abscessus had a 71% inducible resistance rate and a 3% acquired resistance rate, which were comparable to results of a previous study [53]. However, there was a significant difference in susceptibility to clarithromycin between the two MABC species: 94% of M. abscessus subspecies massiliense were susceptible to clarithromycin, in contrast to a low susceptibility to the drug for M. abscessus subspecies abscessus (p<0.001), as observed in other studies [49,53]. The status of erm(41) gene functionality can lead to this difference in macrolide susceptibility [53,54]. These results highlight that correct identification of NTM infection is of utmost importance to guide an appropriate treatment using species-specific strategies.
Our study has several limitations. Firstly, we did not investigate the correlation of positive culture isolate with clinical or radiologic findings mainly due to the retrospective nature of this study. Given the copious number of clinical specimens, manual analysis of clinical features and radiologic findings was not possible. Thus, caution should be exercised when performing data interpretation because NTM isolation rate does not necessarily reflect the rate of NTM clinical infections. In addition, more than one spelight on the clinical significance of
cimen per patient were included in this study. Therefore, the recovery rate of NTM could be biased by this type of sampling method. Secondly, out of positive cultures for NTM, only a small portion (around 30%) were tested for species identification. In this regard, results should be interpreted with caution. Thirdly, as a tertiary hospital, most of our patients and samples came from a single province of the country. Thus, findings of this study might not be generalized to other regions or countries. In addition, the correlation between in vitro AST and in vivo clinical features was not studied, as stated previously. Future studies using patient outcomes are needed to shed light on the clinical significance of in vitro NTM AST in our hospital. Lastly, proper AST results for specimens with multispecies infection were not accomplished. Since multispecies NTM infection in a patient is not an uncommon finding, accurate AST technique using single culture isolate is necessary in the case of multispecies infection.
In summary, the recovery rate of NTM showed an increasing trend in our hospital, although the trend was not observed in 2020. It would be interesting to determine whether the COVID-19 pandemic and crisis-related regulations play a role in the change of epidemiology of NTM infection. MAC, M. abscessus subspecies abscessus, and M. kansasii were the most common NTM species in our center. Little less than 9% of total isolates were shown to be mixed NTM infection. Most of isolated NTM species were susceptible to key drugs such as clarithromycin, amikacin, and rifampin. However, most of M. abscessus subspecies abscessus isolates showed inducible resistance to clarithromycin.
Notes
Supplementary Material
Supplementary material can be found in the journal homepage (http://www.e-trd.org).
Supplementary Table S1. Antimycobacterial agents and breakpoints for testing nontuberculous mycobacteria.
Supplementary Table S1.
Antimycobacterial agents and breakpoints for testing nontuberculous mycobacteria
Fig. 1.
Annual isolation rate of nontuberculous mycobacteria among total clinical specimens compared to Mycobacterium tuberculosis.
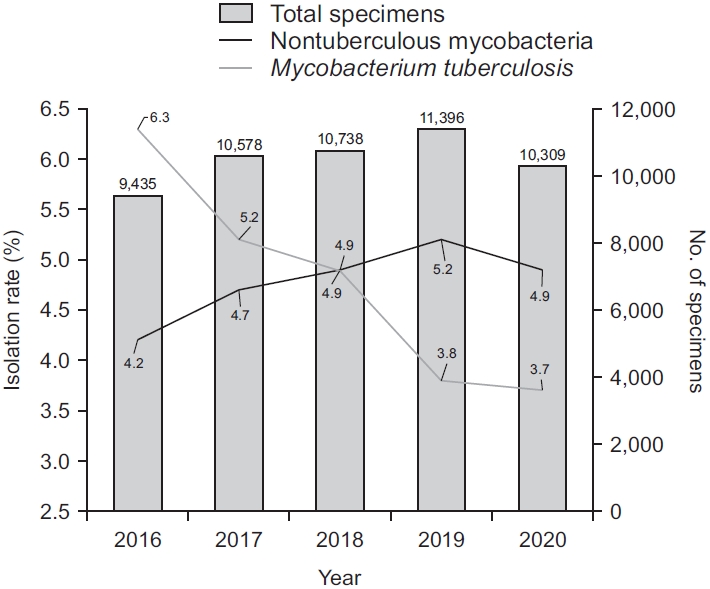
Fig. 3.
Annual proportion of nontuberculous mycobacteria (NTM) species identification (n=718). M. massiliense: Mycobacterium abscessus subspecies massiliense; M. abscessus: Mycobacterium abscessus subspecies abscessus.
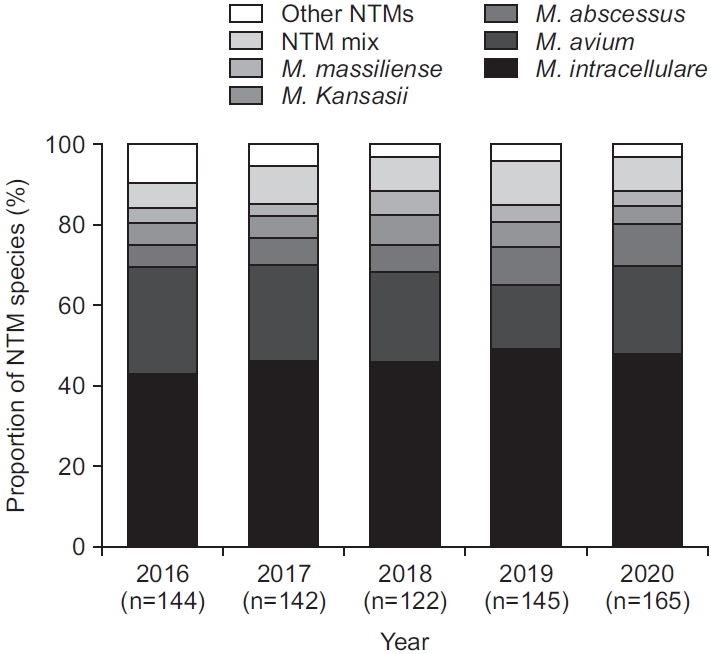
Table 1.
List of identified nontuberculous mycobacteria species recovered from clinical isolates during 2016 to 2020 (n=718)
Table 2.
Antimicrobial susceptibility testing results for Mycobacterium avium complex
Table 3.
Antimicrobial susceptibility testing results for Mycobacterium kansasii
Table 4.
Antimicrobial susceptibility testing results for Mycobacterium abscessus complex
REFERENCES
1. Jeon D. Infection source and epidemiology of nontuberculous mycobacterial lung disease. Tuberc Respir Dis (Seoul) 2019;82:94-101.



2. Kim HS, Lee Y, Lee S, Kim YA, Sun YK. Recent trends in clinically significant nontuberculous Mycobacteria isolates at a Korean general hospital. Ann Lab Med 2014;34:56-9.


3. Kim N, Yi J, Chang CL. Recovery rates of non-tuberculous mycobacteria from clinical specimens are increasing in Korean tertiary-care hospitals. J Korean Med Sci 2017;32:1263-7.




4. Cowman S, Burns K, Benson S, Wilson R, Loebinger MR. The antimicrobial susceptibility of non-tuberculous mycobacteria. J Infect 2016;72:324-31.


5. Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med 2012;185:881-6.



6. Martin-Casabona N, Bahrmand AR, Bennedsen J, Thomsen VO, Curcio M, Fauville-Dufaux M, et al. Non-tuberculous mycobacteria: patterns of isolation: a multi-country retrospective survey. Int J Tuberc Lung Dis 2004;8:1186-93.

7. Shah NM, Davidson JA, Anderson LF, Lalor MK, Kim J, Thomas HL, et al. Pulmonary Mycobacterium avium-intracellulare is the main driver of the rise in non-tuberculous mycobacteria incidence in England, Wales and Northern Ireland, 2007-2012. BMC Infect Dis 2016;16:195.




8. Johansen MD, Herrmann JL, Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 2020;18:392-407.



9. Saxena S, Spaink HP, Forn-Cuni G. Drug resistance in nontuberculous mycobacteria: mechanisms and models. Biology (Basel) 2021;10:96.



10. Sim YS, Park HY, Jeon K, Suh GY, Kwon OJ, Koh WJ. Standardized combination antibiotic treatment of Mycobacterium avium complex lung disease. Yonsei Med J 2010;51:888-94.



11. Wallace RJ, Dukart G, Brown-Elliott BA, Griffith DE, Scerpella EG, Marshall B. Clinical experience in 52 patients with tigecycline-containing regimens for salvage treatment of Mycobacterium abscessus and Mycobacterium chelonae infections. J Antimicrob Chemother 2014;69:1945-53.



12. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 2007;175:367-416.


13. Mirsaeidi M, Farshidpour M, Allen MB, Ebrahimi G, Falkinham JO. Highlight on advances in nontuberculous mycobacterial disease in North America. Biomed Res Int 2014;2014:919474.




14. van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat 2012;15:149-61.


15. Research Committee of the British Thoracic Society. First randomised trial of treatments for pulmonary disease caused by M avium intracellulare, M malmoense, and M xenopi in HIV negative patients: rifampicin, ethambutol and isoniazid versus rifampicin and ethambutol. Thorax 2001;56:167-72.



16. Woods GL. Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes: CLSI supplement M62. 1st ed. Wayne: Clinical and Laboratory Standards Institute; 2018.
17. Griffith DE, Aksamit TR. Managing Mycobacterium avium complex lung disease with a little help from my friend. Chest 2021;159:1372-81.


18. Kwon BS, Kim MN, Sung H, Koh Y, Kim WS, Song JW, et al. In vitro MIC values of rifampin and ethambutol and treatment outcome in Mycobacterium avium complex lung disease. Antimicrob Agents Chemother 2018;62:e00491-18.




19. Moon SM, Kim SY, Kim DH, Huh HJ, Lee NY, Jhun BW. Relationship between resistance to ethambutol and rifampin and clinical outcomes in Mycobacterium avium complex pulmonary disease. Antimicrob Agents Chemother 2022;66:e0202721.



20. Woods GL. Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes: CLSI standard M24. 3rd ed. Wayne: Clinical and Laboratory Standards Institute; 2018.
21. Park Y, Park YE, Jhun BW, Park J, Kwak N, Jo KW, et al. Impact of susceptibility to injectable antibiotics on the treatment outcomes of Mycobacterium abscessus pulmonary disease. Open Forum Infect Dis 2021;8:ofab215.




22. Basille D, Jounieaux V, Andrejak C. Treatment of other nontuberculous mycobacteria. Semin Respir Crit Care Med 2018;39:377-82.


23. Global Laboratory Initiative. Mycobacteriology laboratory manual. 1st ed. Geneva: Global Laboratory Initiative; 2014.
24. Kim SH, Shin JH. Identification of nontuberculous mycobacteria from clinical isolates and specimens using AdvanSure Mycobacteria GenoBlot assay. Jpn J Infect Dis 2020;73:278-81.


25. Petti CA, Brandt ME, Church DL, Emler S, Simmon K, Zelazny AM. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing: CLSI guideline MM18. 2nd ed. Wayne: Clinical and Laboratory Standards Institute; 2018.
26. Yoo JW, Jo KW, Kim MN, Lee SD, Kim WS, Kim DS, et al. Increasing trend of isolation of non-tuberculous mycobacteria in a tertiary university hospital in South Korea. Tuberc Respir Dis (Seoul) 2012;72:409-15.



27. Park Y, Kwak SH, Yong SH, Lee SH, Leem AY, Kim SY, et al. The association between behavioral risk factors and nontuberculous mycobacterial pulmonary disease. Yonsei Med J 2021;62:702-7.




29. Amar S, Avni YS, O’Rourke N, Michael T. Prevalence of common infectious diseases after COVID-19 vaccination and easing of pandemic restrictions in Israel. JAMA Netw Open 2022;5:e2146175.



30. Park YS, Lee CH, Lee SM, Yang SC, Yoo CG, Kim YW, et al. Rapid increase of non-tuberculous mycobacterial lung diseases at a tertiary referral hospital in South Korea. Int J Tuberc Lung Dis 2010;14:1069-71.

31. Spaulding AB, Lai YL, Zelazny AM, Olivier KN, Kadri SS, Prevots DR, et al. Geographic distribution of nontuberculous mycobacterial species identified among clinical isolates in the United States, 2009-2013. Ann Am Thorac Soc 2017;14:1655-61.



32. Lim AY, Chotirmall SH, Fok ET, Verma A, De PP, Goh SK, et al. Profiling non-tuberculous mycobacteria in an Asian setting: characteristics and clinical outcomes of hospitalized patients in Singapore. BMC Pulm Med 2018;18:85.




33. Wu J, Zhang Y, Li J, Lin S, Wang L, Jiang Y, et al. Increase in nontuberculous mycobacteria isolated in Shanghai, China: results from a population-based study. PLoS One 2014;9:e109736.



34. Honda JR, Virdi R, Chan ED. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front Microbiol 2018;9:2029.



35. Zhang H, Luo M, Zhang K, Yang X, Hu K, Fu Z, et al. Species identification and antimicrobial susceptibility testing of non-tuberculous mycobacteria isolated in Chongqing, Southwest China. Epidemiol Infect 2020;149:e7.



36. Zhang ZX, Cherng BP, Sng LH, Tan YE. Clinical and microbiological characteristics of non-tuberculous mycobacteria diseases in Singapore with a focus on pulmonary disease, 2012-2016. BMC Infect Dis 2019;19:436.




37. Khieu V, Ananta P, Kaewprasert O, Laohaviroj M, Namwat W, Faksri K. Whole-genome sequencing analysis to identify infection with multiple species of nontuberculous mycobacteria. Pathogens 2021;10:879.



38. Woodley CL, Kilburn JO, David HL, Silcox VA. Susceptibility of mycobacteria to rifampin. Antimicrob Agents Chemother 1972;2:245-9.




39. Rosenzweig DY. Pulmonary mycobacterial infections due to Mycobacterium intracellulare-avium complex: clinical features and course in 100 consecutive cases. Chest 1979;75:115-9.


40. Etzkorn ET, Aldarondo S, McAllister CK, Matthews J, Ognibene AJ. Medical therapy of Mycobacterium avium-intracellulare pulmonary disease. Am Rev Respir Dis 1986;134:442-5.

41. Wallace RJ, Dunbar D, Brown BA, Onyi G, Dunlap R, Ahn CH, et al. Rifampin-resistant Mycobacterium kansasii. Clin Infect Dis 1994;18:736-43.


42. Griffith DE, Brown-Elliott BA, Langsjoen B, Zhang Y, Pan X, Girard W, et al. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 2006;174:928-34.


43. Brown-Elliott BA, Iakhiaeva E, Griffith DE, Woods GL, Stout JE, Wolfe CR, et al. In vitro activity of amikacin against isolates of Mycobacterium avium complex with proposed MIC breakpoints and finding of a 16S rRNA gene mutation in treated isolates. J Clin Microbiol 2013;51:3389-94.




44. Jeon K, Kwon OJ, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med 2009;180:896-902.


45. Koh WJ, Hong G, Kim SY, Jeong BH, Park HY, Jeon K, et al. Treatment of refractory Mycobacterium avium complex lung disease with a moxifloxacin-containing regimen. Antimicrob Agents Chemother 2013;57:2281-5.




46. Brown-Elliott BA, Woods GL. Antimycobacterial susceptibility testing of nontuberculous mycobacteria. J Clin Microbiol 2019;57:e00834. -19.




47. Jaffre J, Aubry A, Maitre T, Morel F, Brossier F, Robert J, et al. Rational choice of antibiotics and media for Mycobacterium avium complex drug susceptibility testing. Front Microbiol 2020;11:81.



48. Huang WC, Yu MC, Huang YW. Identification and drug susceptibility testing for nontuberculous mycobacteria. J Formos Med Assoc 2020;119 Suppl 1:S32-41.


49. Liu CF, Song YM, He WC, Liu DX, He P, Bao JJ, et al. Nontuberculous mycobacteria in China: incidence and antimicrobial resistance spectrum from a nationwide survey. Infect Dis Poverty 2021;10:59.




50. Cho EH, Huh HJ, Song DJ, Moon SM, Lee SH, Shin SY, et al. Differences in drug susceptibility pattern between Mycobacterium avium and Mycobacterium intracellulare isolated in respiratory specimens. J Infect Chemother 2018;24:315-8.


51. Moon SM, Choe J, Jhun BW, Jeon K, Kwon OJ, Huh HJ, et al. Treatment with a macrolide-containing regimen for Mycobacterium kansasii pulmonary disease. Respir Med 2019;148:37-42.


52. da Silva Telles MA, Chimara E, Ferrazoli L, Riley LW. Mycobacterium kansasii: antibiotic susceptibility and PCR-restriction analysis of clinical isolates. J Med Microbiol 2005;54((Pt 10):975-9.

- TOOLS
-
METRICS

- ORCID iDs
-
Keun Ju Kim

https://orcid.org/0000-0003-2548-5609Chulhun L. Chang

https://orcid.org/0000-0001-9117-4919 - Related articles


 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Data Sharing Statement
Data Sharing Statement Full text via DOI
Full text via DOI Supplement
Supplement Print
Print Download Citation
Download Citation