Year-in-Review of Lung Cancer
Article information
Abstract
In the last several years, we have made slow but steady progress in understanding molecular biology of lung cancer. This review is focused on advances in understanding the biology of lung cancer that have led to proof of concept studies on new therapeutic approaches. The three selected topics include genetics, epigenetics and non-coding RNA. This new information represents progress in the integration of molecular mechanisms that to identify more effective ways to target lung cancer.
Introduction
Lung cancer is the leading cause of cancer-related mortality in the developed world, accounting for 26~29% of all cancer deaths1. Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancer diagnosis and the majority (60~80%) of patients are diagnosed at an advanced stage. Consequently, the prognosis for NSCLC remains poor, with a 5-year survival rate of 15%2.
The histologic subtype of lung cancer has had little impact on the selection of therapy. Non-squamous cell histology is an indication for chemotherapy such as pemetrexed and bevacizumab. This type of stratification represents the first step toward personalized therapy in NSCLC3. Beyond histology, some molecular alterations have been shown to correlate with response to treatment with chemotherapy. New therapies targeting the EML4-ALK, BRAF, FGFR and other molecular alterations are under way to help define specific subsets of patients responsive to certain molecularly targeted treatments4.
The ongoing identification of novel molecular abnormalities in lung cancer continues to present exciting opportunities in targeted therapy. In this brief review, there will be an effort to discuss some of the developments in lung cancer biology. As an in depth review of all areas of biology is not possible within this manuscript, three fields (genetics, epigenetics and non-coding RNA) were selected for detailed review (Figure 1).
Literature Search
A literature search from January to December 2011 was conducted using PubMed and the following search terms: "Non-small cell lung cancer/molecular biology," "Non-small cell lung cancer/genetics," "Non-small cell lung cancer/epigenetics" and "Lung cancer/non-coding RNA." Conference proceedings in 2011 for American Society of Clinical Oncology (ASCO), American Association for Cancer Research (AACR), and the International Society for the Study of Lung Cancer (IASLC) World Lung Congresses were searched.
Genetics
Analysis of cancer genome sequences using DNA microarrays and capillary-based DNA sequencing provide insights for understanding cancer biology, diagnosis and therapy. However, there are particular challenges for the detection and diagnosis of cancer genome alterations. For example, some genomic alterations in cancer are prevalent at a low frequency in clinical samples, often owing to substantial admixture with non-malignant cells. Next-generation sequencing technologies can solve such problems. Furthermore, these new sequencing methods make it feasible to discover novel chromosomal rearrangements and microbial infections, and to resolve copy number alterations at very high resolution5.
Several recent studies using next generation sequencing technologies have been reported in lung cancer. Lipson et al.6 analyzed genomic DNA from 24 NSCLC formalin-fixed paraffin-embedded specimens using an assay that captured and sequenced 2,574 coding exons representing 145 cancer-relevant genes (genes associated with cancer-related pathways, targeted therapy or prognosis) plus 37 introns from 14 genes that are frequently rearranged in cancer. Among the 24 NSCLCs, 50 alterations were identified in 21 genes, with at least one alteration being present in 83% (20 out of 24) of the tumors. Twelve genes were altered in multiple tumors. In addition to the known NSCLC gene alterations, G1849T (V617F) JAK2 mutation and gene fusion joining exons 1~15 of KIF5B to exons 12~20 of RET were notable discoveries. KIF5B-RET fusion was also reported in lung adenocarcinoma of Korean patients using next-generation sequencing technologies7. These findings suggest that RET kinase inhibitors and JAK inhibitors should be tested in prospective clinical trials for therapeutic benefit in individuals with NSCLC that carry these genomic alterations. In another novel finding using next generation sequencing technique, Jung et al.8 analyzed the transcriptome of the NSCLC cell line H2228 and discovered a fusion transcript composed of multiple exons of ALK and PTPN3. Detailed analysis of the genomic structure revealed that a portion of genomic region encompassing exons 10 and 11 of ALK had been translocated into the intronic region between exons 2 and 3 of PTPN38. In lung cancer, five fusion partners of ALK have been reported EML4, TFG, KIF5B, KLC1 and PTPN39. As more ALK fusion partners are identified, the population of patients with ALK fusion who may potentially benefit from ALK inhibitor therapy may be extended.
The next-generation sequencing could provide innovative information in cancer biology. But, to be widely and routinely used in clinical practice, there needs to be further reduction in cost, flexibility in throughput and efficiency of data generation, analysis and management.
Epigenetics
Epigenetics is defined as heritable changes in gene expression that are not attributable to changes in the sequence of DNA. Epigenetic regulation of gene expression has emerged as a fundamental pathway in the pathogenesis of lung cancer. The predominant epigenetic mechanisms are DNA methylation and histone modifications10-12. DNA methylation is the covalent addition or subtraction of a methyl group to a cytosine nucleotide in a sequence of DNA. Methylation is controlled by a family of specific enzymes known as DNA methyltransferases (DNMTs). The addition of methyl groups can be highly specific to a particular gene. Hypermethylation of CpG islands in the promoter region of a gene can result in transcriptional silencing of the gene, and subsequent loss of protein expression13. The molecular mechanism of demethylated DNA has long been unknown. The recent discovery that the three members of the ten-eleven translocation (TET) protein family can convert 5-methylcytosine (5-mC) into 5-hydroxymethylcytosine (5-hmC) has provided a potential mechanism leading to DNA demethylation (Figure 2). Moreover, the demonstration that TET2 is frequently mutated in hematopoietic tumors suggests that the TET proteins could play a crucial role in tumorigenesis14. The levels of 5-hmC of lung cancer tissues have been shown to be markedly depleted with up to a 5-fold reduction compared with normal lung tissue15. The biological properties of DNA demethylation by TET proteins in solid tumors was reported earlier this year. Kudo et al.16 reported that down-regulation of TET1 and loss of 5-hmC were induced by oncogene-dependent cellular transformation. These results suggest critical roles of aberrant DNA demethylation for oncogenic processes in solid tissues.
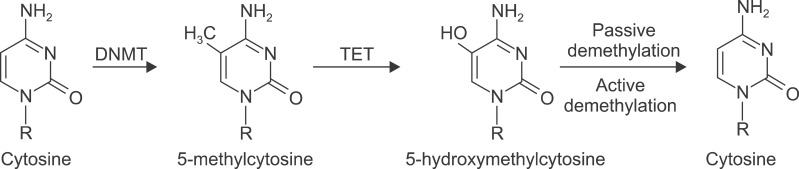
Enzymatic conversion of cytosine to 5-hydroxymethylcytosine. DNA methyltransferases (DNMTs) convert cytosine to 5-methylcytosine. 5-hydroxymethylcytosine be converted to 5-hydroxymethylcytosine in an enzymatic process involving members of the ten-eleven translocation (TET) protein family. 5-hydroxymethylcytosine could be converted to cytosine through a passive demethylation or an active demethylation pathway.
The discovery of 5-hmC and TET in lung cancer has added a new and potentially important dimension to our perception of DNA methylation. Further studies are required to confirm role of 5-hmc and TET and to investigate potential clinical applications in lung cancer.
Non-coding RNAs (ncRNA)
A ncRNA is a functional RNA molecule that is not translated into a protein. ncRNAs are regarded as regulators of cell cycle progression, proliferation, and fate. There are numerous classes of ncRNAs, including microRNAs (miRNA, ~22 nt) and Piwi-interacting RNAs (piRNAs, 18~30 nt), short translational-regulatory RNAs (100~200 nt), and much longer ncRNAs (lncRNAs, up to 10,000 nt)17,18. So far, the function of miRNAs as key components of the RNA interference (RNAi) pathway, their role as tumor suppressors, and their impact on tumorigenesis have been demonstrated. Nevertheless, little is known about lncRNAs and their impact on tumorigenesis regulatory processes. LncRNAs participate in a wide-range of biological processes. Almost every step in the life cycle of genes can be influenced by lncRNAs through posttranscriptional gene regulation, RNA maturation, RNA transport and transcriptional gene silencing. The molecular mechanisms of lncRNAs are classified into four archetypes: signals, decoys, guides and scaffolds (Figure 3). Several lncRNAs possess characteristics from multiple archetypes that, in combination, are critical to their eventual biological function19. Metastasis-associated-in-lung-adenocarcinoma-transcript-1 (MALAT-1) is an lncRNA and highly expressed in several tumor types. MALAT-1 strongly associates with serine-arginine rich splicing factor (SR) proteins involved in both constitutive and alternative splicing, and the levels of MALAT-1 regulate the cellular levels of phosphorylated SR proteins. These findings imply that the MALAT-1 may serve a function in the regulation of alternate splicing by modulating the activity of SR proteins20. MALAT-1 is highly expressed in several human NSCLCs and is a strong regulator of NSCLC migration and invasion. MALAT-1 RNA expression in squamous cell carcinoma of the lung has been associated with a poor prognosis21.

Schematic diagram of the four archetypes of longer ncRNAs (lncRNA) mechanism. Signaling: LncRNA expression can be stimulated in response to certain stimuli, such as cellular stress and temperature. Decoys: Specific lncRNAs are transcribed and then bind to and titrate away protein factors. Guides: LncRNAs can be molecular guides by localizing particular ribonucleoprotein complexes to specific chromatin targets. Scaffolds: LncRNAs can bring together multiple proteins to form ribonucleoprotein complexes. Adopted from Wang and Chang19. Mol Cell 2011;43:904-14.
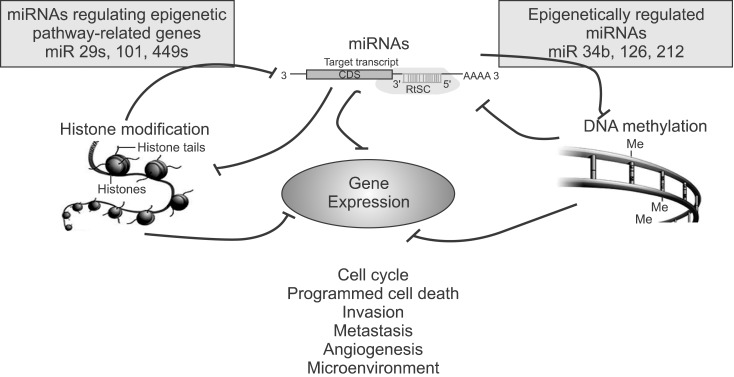
miRNAs regulate gene expression at the post-transcriptional level through sequence-specific interactions with 3'-untranslated regions in mRNAs and also via translation inhibition or degradation of mRNAs. Recent studies have demonstrated that epigenetic mechanisms regulate the expression of miRNAs. An extensive analysis of genomic sequences of miRNA genes has shown that approximately half are associated with CpG islands. Therefore three main epigenetic events including the aberrant hypermethylation of tumor suppressor genes, global DNA hypomethylation and post-translational modifications of histone could affect miRNA expression as well22. Some miRNAs are up-regulated upon the exposure of cells to the agent 5-aza-2'-deoxycytidine, upon mutation of DNMTs or upon treatment with histone deacetylase inhibitors23-25. Conversely, another subset of miRNAs controls the expression of important epigenetic regulators including DNA methyltransferases, histone deacetylases and polycomb group genes26-28. This complicated network of feedback between miRNAs and epigenetic pathways appears to form an epigenetics-miRNA regulatory circuit, which may organize the whole gene expression profile in lung cancer (Figure 4).
It has become evident in recent years that the de-regulation of miRNAs and lncRNAs plays a critical role in malignant transformation and tumor cell behavior. The functional role for the vast majority of these ncRNA is still in question. Further study of ncRNA could yield new RNA-based targets for the prevention and treatment of lung cancer.
Conclusions
Lung cancer is a disease of genetic alteration, epigenetic changes as well as transcription control by non-coding RNA. The biology of lung cancer may also predict response or outcome to certain chemotherapeutic agents, serving as biomarkers that could inform clinical decisions. Along with newly discovered tumor biologic mechanisms, these findings raise hope that cancer can and will be vanquished.
Acknowledgements
The authors thank Jaeku Kang (Department of Pharmacology, College of Medicine, Konyang University) for careful review of the manuscript.